Intro and retrospective on Computational Biology & Data
Live blogging from conference
Data, big data, omics data, noisy data, a lack of data, data overflow, open data.Data is an increasingly critical part of life as a researcher — particularly in the field of computational biology. How to treat data? How to manage data? How to smartly produce data? How to simulate data? How to reproduce data? Data, data, data…
But what is Computational Biology? In the past, it was seen as a support for biological research; a set of tools helping answer biological questions asked by researchers — real researchers that is. However, when you consider recent breakthroughs, dynamical development of this discipline and the impact it has, it is easy to see why those in the field believe that that computational biology is and will be a driving force of biological research. What is particularly striking is the change in computational biology from descriptive to data-driven science.

It all started with genomics. When biologist trained in wet-lab work they could not treat their data themselves and this is how new discipline was born. Like all young disciplines, it had to negotiate its place in the research landscape. Today, computational biology is still expanding thanks to new technologies and the ongoing digital revolution.
Breakthroughs
An undeniable innovator of computational biology, Serafim Batzoglou, in his keynote talk at ISMB 2016, presented his vision of the past and the future of this discipline. His talk started with human genome sequencing, through to data manipulation, to personal genomics and finally artificial intelligence. He stated research need to keep up with new technologies with a quote from the TV series Silicon Valley:
“I don’t want to live in a world where someone else is making the world a better place better than me”
In his talk, Serafim Batzoglou made the somewhat controversial comment that AI will replace medical doctors in near future and he named his candidate for most important and promising technology: deep learning.
Deep learning is a branch of machine learning based on a set of algorithms that attempt to model high-level abstractions in data by using a deep graph with multiple processing layer; composed of multiple linear and non-linear transformations.
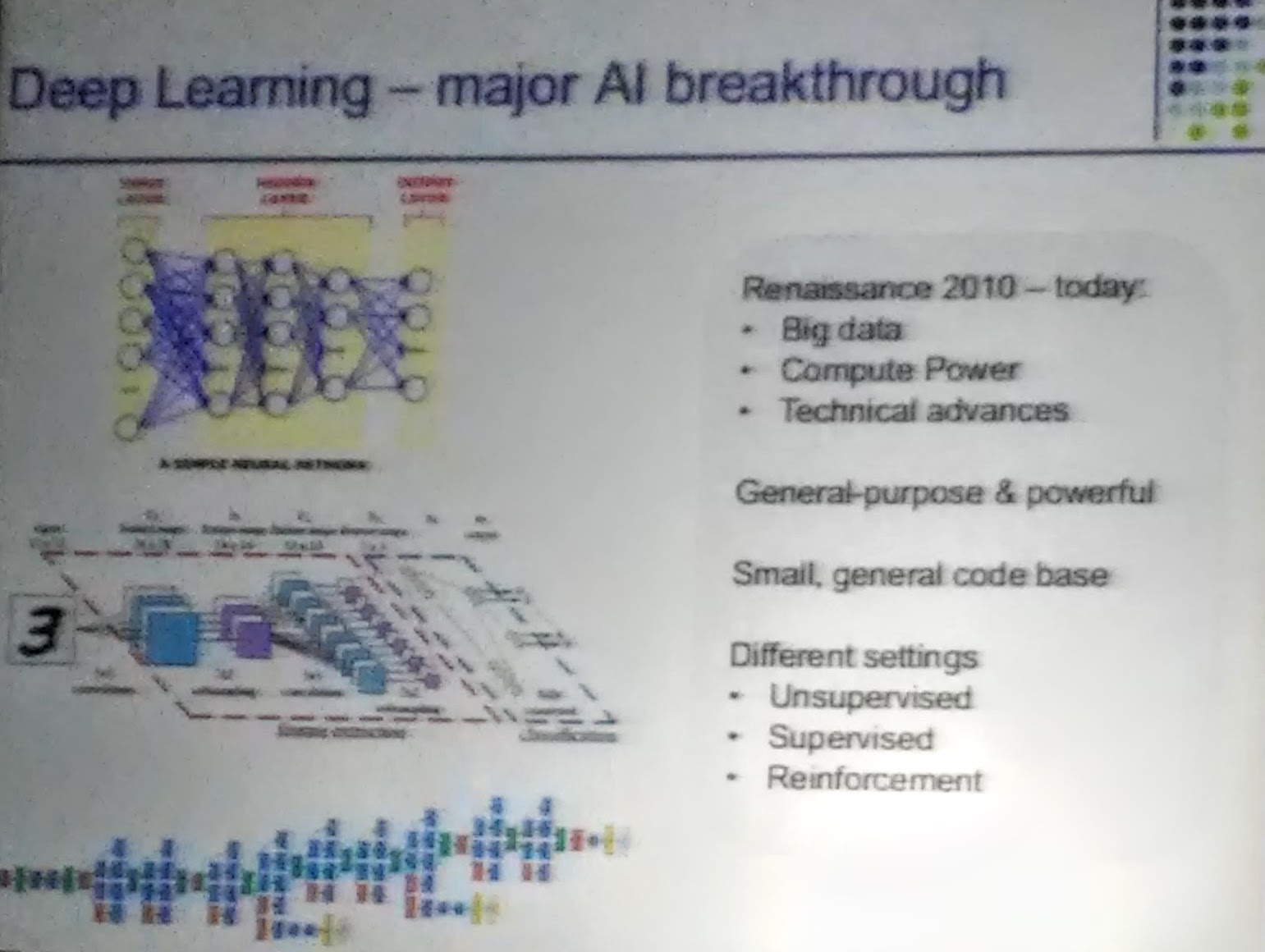
Single-cell transcriptomics were unofficially named most exciting technology for studying biological systems - with three out of six keynote speakers talking about their recent progress when treating single cell transcriptomics data.
Serafim Batzoglou also mentioned clustering algorithm, called SimlR challenging existing methods of dimension reduction . The preprint of the article is accessible at http://biorxiv.org/.
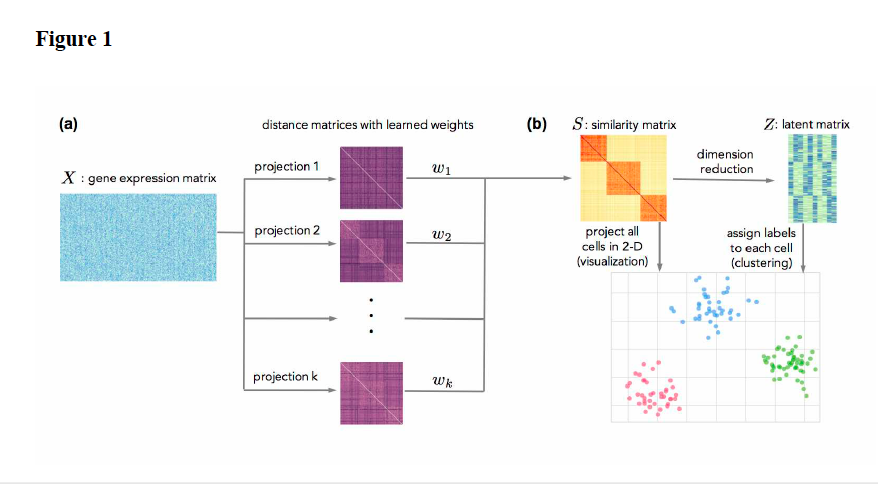
Sandrine Dudoit also referred to single-cell data; with particular attention to treating the problem of noise to signal ratio and data reproducibility. In her lab more robust tools of clustering are under development. For example: clusterExperiment decides on clusters based on many clustering methods and picks the ones that are the most stable.
The keynote talk by Sarah Teichmann showcased a cutting-edge example of exploration of single-cell data. In publication she co-authors (“Tcell fate and clonality inference from single-cell transcriptomes”), full-length paired T-cell receptor (TCR) sequences are reconstructed from single cell data.
A key trend currently is to gather information at the highest level of accuracy and then group it into meaningful blocks of knowledge which best characterize the data and correspond to biological reality, or even better, discover unknown pieces of biological systems. This is how computational biology is driving biological research today.
And there will be more; more data, more challenges, more breathtaking discoveries!
Challenges
One them is education. Phil Bourne, Associate Director for Data Science of NIH, in the workshop Education in Bioinformatics — Exploiting Cloud and Virtual Resources for Training, highlighted the importance of cloud data training to keep the pace of technologies developed by companies (e.g. Amazon, Google, iPlant). The new generation of computational biologists and Bioinformaticians, as well as senior researchers, require solid and up-to-date training. This is why the NIH proposes numerous programs facilitating cloud training. Unfortunately, it is not the case at all research institutes currently, but it definitely should be in the future.
Another important issue raised by Phil Bourne and Serafim Batzoglou was the importance of keeping data open. Phil mentioned in particular the FAIR Data Principles which stand for:
Findability, Accessibility, Interoperability, and Reusability - These serve to guide data producers and publishers as they navigate around obstacles, thereby helping to maximize the added-value gained by contemporary, scholarly publishing.
Serafim Batzoglou outlined the necessity of making data public in one of the slides in his keynote talk (quoted):
Biggest obstacle: wide, free data availability
Ideal
- Millions of publicity available genomes, medical records, phenotypes, enivornments
- Far reaching anti data-based discrimination laws; GINA great start
Not ideal
- Data silos in hospitals, health care providers, gov’t, direct-to-direct consumer companies
- Specialized large-scale projects (1000 genomes but no phenotypes/medical records)
What about privacy?
- No cancer patients have died yet because of data privacy breach
- My Facebook page reveals much more private stuff about me than my genome or medical record
Let’s make our data public — if 5% of us do, probably enough to make a big difference. Let’s do it without waiting for legislation!
Flaws
There are still some flaws with regards to big data — but it still represents big hope for computational biology.
According to Uri Laserson from Cloudera (a leader platform for data management and analysis), “Genomic is not special”. He expressed the view that:
Computational biologists are reinventing the wheel for big data biology analysis
Even though the statement is bold, there is some truth in it. It looks like industrial solutions for big data somehow outperform research solutions. This is why more collaboration between research and industry is needed to fill the gap. On the other hand, this collaboration cannot happen at the price of data ownership. Academy needs to find a solution to attract world-scale talents that right now goes where big data is linked tightly to big money.
Another recurrent problem for many computational biologists and bioinformaticians is their place in biological research. Despite an improving amount of recognition of computational work in wet labs, there is still a number of places that lack of expertise in data analysis. Old analysis methods are reproduced and consequently spread used by others in the field — all because a number of researchers are using them.
Programming or statistical input is not acknowledged enough and this is where there can some friction between experimentalists and theoreticians that can hinder progress. Strolling through alleys of the poster hall at ISMB 2016, overheard many accounts of interactions between wet and dry lab. Most of them recalled tension, disappointment or misunderstanding. This is why education is really crucial. No one cannot be an expert in all domains, but a comprehensible overview of each other worlds, common vocabulary and goals are essential to building a common future.
Computational biology is in the heart of digital revolution. Paradoxically a lack and an excess of data make this discipline extremely intriguing and challenging. With individual countries frequently making decisions on policies that have an impact on the direction and pace of the development of solutions, data’s impact on our future is undeniable. But then again, what is data? A set of numbers?
From technical point of view, each of us is a multidimensional data-set. Can we be easily reduced to a few dimensions? Can our time dimension be predicted from real time data? Can someone own us? Are we approaching era of digital slavery?

You may be also interested in :
Computational Biology: Moving into the Future One Click at a Time. Message from ISCB
Disclaimer: Any views expressed are those of the author, not necessarily those of PLOS.