The Mamba Effect
Mamba models gaining ground in artificial intelligence research.
Already 8 papers since December 2023 !
Mamba is already a new wave starting as a replacement for vanilla transformer, it has already been adapted to text, vison, video…
Mamba models represent a significant breakthrough in neural network architecture.
Among published papers, besides the original Mamba paper, I would like to distinguish two spin-offs: VMamba and MambaBytes. There it also a bunch of papers with specific biomedical applications that I am not expert to evaluate impact.
Here I compiled a short overview of all (?) those papers adapting the template from The Batch newsletter from deeplearning.ai.
here is a list of mamba papers : https://github.com/yyyujintang/Awesome-Mamba-Papers
Arxiv 23.12.01: Mamba: Linear-Time Sequence Modeling with Selective State Spaces
#mamba
This paper is the first to introduce the Mamba architecture. Mamba offers faster inference, linear scaling with sequence length, and strong performance.
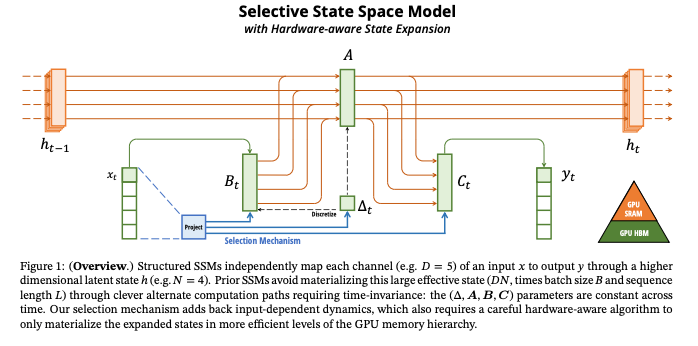
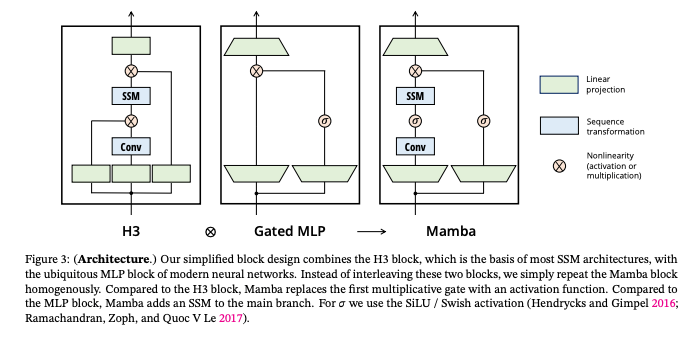
What’s New
- A new architecture, Mamba, integrating SSMs without relying on attention or MLP blocks.
- Implementation of a hardware-aware parallel algorithm for efficient computation.
- Long context: the quality and efficiency together yield performance improvements on real data up to sequence length 1M
How it Works: Mamba adds recurrent and convolutional models, to a unique selection mechanism that enables the model to prioritize or ignore inputs based on the content relevance. This approach allows for linear scalability in sequence length. Mamba integrates selective SSMs into a simplified neural network architecture with gates. They are structured to enable the model to selectively propagate or to forget information based on the current token.
Results: Mamba demonstrates state-of-the-art performance across various modalities. In language modeling, Mamba-3B outperforms Transformers of the same size, matches Transformers twice its size in both pretraining and downstream evaluation. In terms of efficiency, it achieves 5× higher throughput than Transformers and scales linearly with sequence length. This in various domains such as language, genomics, audio modeling. It efficiently handles sequences up to a million lengths.
Behind the News: The development of Mamba marks a significant shift from the dominant Transformer-based architectures. It opens new avenues in sequence modeling, especially for applications requiring efficient processing of long data sequences. Authors mentions their ambition to make Mamba alternative to Tranformers and CNN as a Foundation Model backbone.
Why it Matters: Mamba has the potential to revolutionize various applications in deep learning. This first paper already resulted in 7 other papers in the same month as code is under apache licene and public.
We’re Thinking: We are missing distance to see how Mamba module will perform in practice. However the fact that many publicaitons already applied Mamba, modified it and obtained pulishable resutls is promising.
In Detail:
Selective SSMs

The key contribution of the paper lies in the novel implementation of Selective State Space Models. It leverages parameters that control if the model response to current inputs or it maintains its existing state. For instance, a parameter ∆ (Delta) in the model’s architecture determines the balance between focusing on the current input (larger ∆ values) and preserving the ongoing state (smaller ∆ values). The selective modulation of parameters B and C tunes how the inputs influence the state and, conversely, how the state influences the outputs. The selective approach also manages context and resets boundaries in scenarios where sequences are concatenated. This prevents the unwanted bleed of information between concatenated sequences.
Other ressources:
- building-mamba-from-scratch-a-comprehensive-code-walkthrough
- mamba-revolutionizing-sequence-modeling-with-selective-state-spaces
- mamba-and-state-space-models-explained
Arxiv 24.01.08: MoE-Mamba: Efficient Selective State Space Models with Mixture of Experts
#LLM
The paper introduces a novel model in the field of sequential modeling.
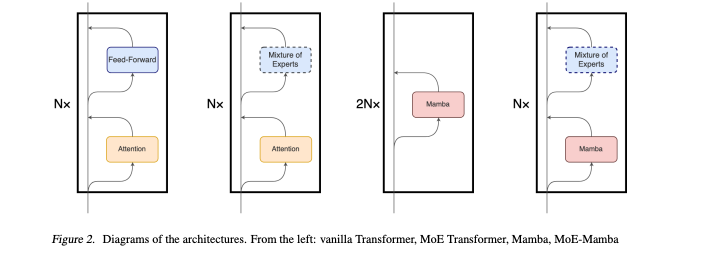
Introduction: The paper presents MoE-Mamba, a model that integrates State Space Models (SSMs) with Mixture of Experts (MoE) to enhance sequential modeling. This combination aims to leverage the strengths of both SSMs, known for their efficient performance, and MoE, a technique for scaling up models efficiently.
What’s New: Integration of SSMs and MoE. MoE layers are used by Mistral, one of SOTA LLM models.
How it Works: The MoE-Mamba architecture replaces every other Mamba layer with a MoE feed-forward layer.
Results - MoE-Mamba achieves better performance than both Mamba and Transformer-MoE models. - It reaches the same performance as Mamba in significantly fewer training steps. - The model scales well with the number of experts, with optimal results at 32 experts.
Behind the News: The scalability potential of MoE-Mamba is remarkable. Although the current study focuses on smaller models, the architecture suggests a promising avenue for handling larger, more complex models efficiently.
Why it Matters: Given the efficiency in training and inference, MoE-Mamba shows promise for deployment in large-scale language models.
We’re Thinking: It is also not clear what are the scalability limits of MoE-Mamba, especially in comparison to existing large-scale models like GPT-3? Also, we would like to have a look a the code…
In Detail
Integration of MoE layers into the Mamba architecture
This design choice enables MoE-Mamba to leverage the conditional processing capabilities of MoE and the context integration of Mamba. By alternating between unconditional processing by the Mamba layer and conditional processing by a MoE layer, MoE-Mamba achieves a balance between efficient state compression and selective information retention.
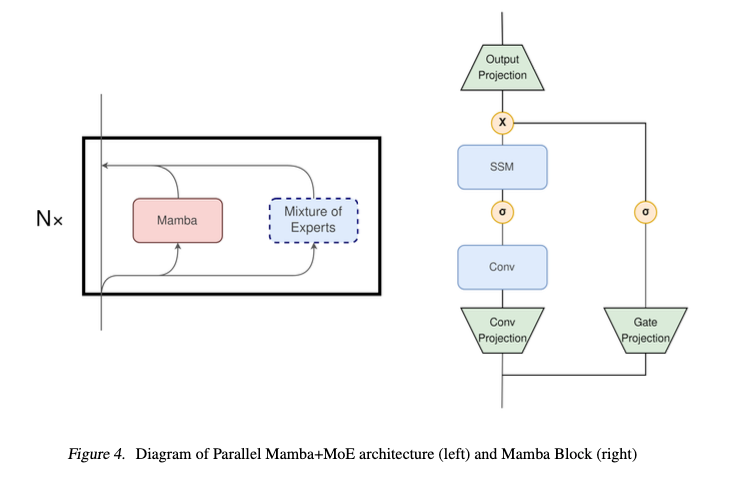
Authors also investigate a unified Mamba module containg MoE
Arxiv 24.01.09: U-Mamba: Enhancing Long-range Dependency for Biomedical Image Segmentation
#biomedical #segmentation #cv
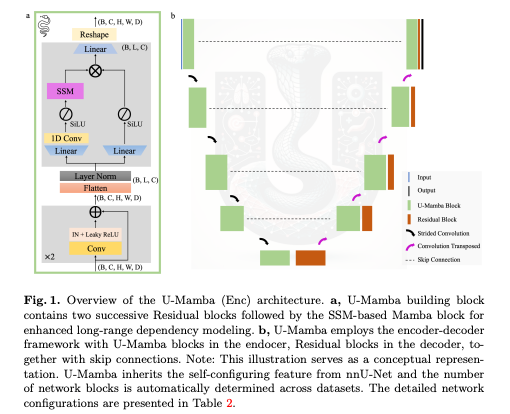
This paper presents an innovative network architecture for biomedical image segmentation.
What’s new: U-Mamba is a novel network integrating Mamba blocks, into a U-Net based architecture. This hybrid CNN-SSM structure enables modeling of long-range dependencies in images.
How it works: U-Mamba follows an encoder-decoder structure, where each building block comprises Residual blocks followed by a Mamba block. This design captures both local features and long-range dependencies. The network’s self-configuring mechanism allows it to adapt automatically to various datasets.
Results: The experiments across biomedical segmentation tasks show that U-Mamba outperforms the state-of-the-art CNN/Transformer-based networks in terms of segmentation accuracy by a small margin.
Behind the news: The integration of Mamba blocks within a U-Net architecture represents an interesting integration of two architectures, highlighting the potential of State Space Models in this domain.
Why it matters: Yet one more architecture smoothly integrating Mamba with good results.
We’re thinking: The U-Mamba network’s adaptability and performance set a new benchmark in medical image segmentation. This work might stimulate further exploration of hybrid architectures in medical image analysis.
While U-Mamba shows some advantages over existing methods, the improvement margin in some cases appears modest. For example, in 3D organ segmentation, U-Mamba’s DSC scores are marginally higher than nnU-Net, which is one of the closest competitors.
Authors do not support with numbers efficiency of the model, model size and comparison in that matter of the two architecture variants.
Should we wait for U-Mamba perfomance for standart segmentation benchmarks such as ADE20K or PASCAL to decide U-Mamba generic value?
In Detail
U-Mamba Architecture
The U-Mamba architecture follows the encoder-decoder pattern of U-Net, known for its effectiveness in medical imaging.
Building Blocks: Each block contains two successive Residual blocks followed by a Mamba block. The Residual block includes a plain convolutional layer, Instance Normalization, and Leaky ReLU.
Mamba Block: It processes image features that are flattened and transposed, followed by Layer Normalization. The Mamba block has two parallel branches:
- The first branch expands the features and processes them through a linear layer, a 1D convolutional layer, SiLU activation, and the SSM layer.
- The second branch also expands the features, followed by SiLU activation. Features from both branches are then merged using the Hadamard product and projected back to their original shape.
Encoder and Decoder: The U-Mamba encoder, captures both local features and long-range dependencies. The decoder focuses on local information and resolution recovery, using Residual blocks, transposed convolutions, and inherits skip connections from U-Net. The output is passed through a convolutional layer and a Softmax layer for the final segmentation probability map.
Variants:
- U-Mamba_Bot: Uses the U-Mamba block only in the bottleneck.
- U-Mamba_Enc: Employs the U-Mamba block in all encoder blocks.
Arxiv 24.01.17: Vision Mamba: Efficient Visual Representation Learning with Bidirectional State Space Model
#computervision #classification #segmentation
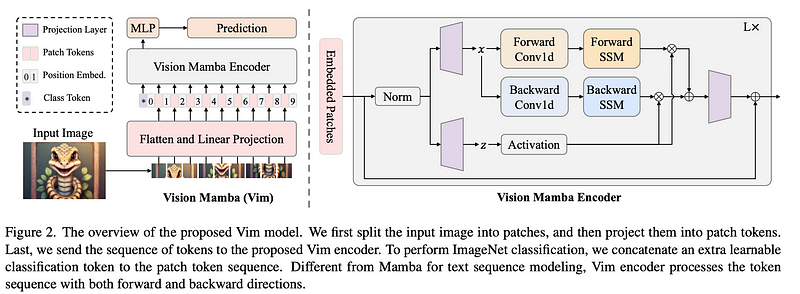
Vim utilizes bidirectional State Space Models (SSMs) to process image sequences.
What’s New
- Vim proposes a pure-SSM-based method for vision tasks, differing from self-attention-based models.
- Incorporation of bidirectional SSMs for efficient visual data processing.
How It Works
- Vim transforms images into sequences of flattened 2-D patches, applying bidirectional SSM.
- The system uses a combination of position embeddings and bidirectional state space models for visual data processing.
Results
- On tasks like ImageNet classification Vim outperforms established models like DeiT and ViTs in terms of accuracy with smaller size.
- Authors compare also segmentation UperNet framework with Vim, DeiT and ResNet on ADE20k with similar conclusion.
- For object detection benchmark Vim slighly outperforms DeiT in the scope of Cascade Mask R-CNN on COCO.
Behind the News: Vim challenges the dominance of self-attention in visual representation, offering an alternative that’s more efficient in handling large-scale and high-resolution datasets.
Why It Matters: Vim’s efficiency in processing high-resolution images makes it a promising backbone for future vision foundation models.
We’re Thinking: Will Vim show its efficiency/accuracy across other frameworks than UperNet or Cascade Mask R-CNN ?
In Detail
Bidirectional State Space Model
- It processes the visual data both forward and backward, unlike traditional unidirectional models.
- This bidirectionality allows for more robust capturing of visual contexts and dependencies, particularly in dense prediction tasks.
- The model effectively compresses the visual representation, leveraging position embeddings to maintain spatial awareness.
Arxiv 24.01.18 : VMamba: Visual State Space Model
#computervision #classification #segmentation #detection
VMamba presents new approach in visual representation learning. It merges CNNs’ and ViTs’ strengths and does not have their limitations.
What’s new: VMamba has a unique architecture: integrates global receptive fields and dynamic weights within a linear computational complexity framework.
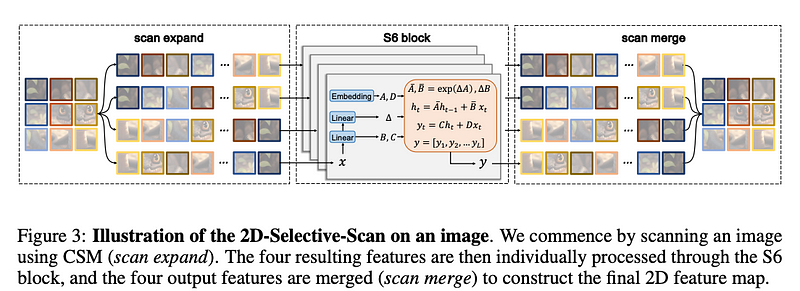
How it works: The model is based on a Cross-Scan Module (CSM). This module processes visual data in four directions. This ensures global information integration without increased complexity. The 2D Selective Scan combines CMS with S6 mamba block and merge output features creating 2D feature map.
Results: VMamba shows significant performance improvements in different task : image classification, object detection, and semantic segmentation. For instance, it surpasses established benchmarks like ResNet and Swin in ImageNet-1K classification. In COCO object detection, VMamba models outperform their counterparts in mean Average Precision (mAP) and mean Intersection over Union (mIoU).
Behind the news: VMamba maintains high performance across various input image sizes, which can indicate a robustness to changes in input conditions. This feature is crucial for practical applications where image resolutions can vary significantly.
It dethrones Vim just the next day of Vim publication.
Why it matters: VMamba’s approach combines the strengths of CNNs and ViTs and on the top it is computationally efficient.

We’re thinking: While VMamba’s results are promising, its practical applicability in diverse real-world scenarios require time verification. For data scientists, VMamba holds a promise of efficient processing of large-scale image datasets and a new playground.
In Detail
VMamba’s 2D Selective Scan operates by dynamically adjusting weights based on the importance of different areas in an image. This process involves an algorithm that assesses each pixel’s contribution to the target task. The scan prioritizes regions with higher information content, therefore it reduces computational load on less relevant areas. This method contrasts with traditional approaches where all pixels are treated equally. Which also leads to higher computational costs.

Vim vs VMamba
VMamba beats Vim on ImageNet-1k benchmark
- VMamba-T with 22M params achieves 82.2 % acc while comparable VimS with 26M achieves 80.3% acc
VMamba beats Vim on COCO benchmark
- the smalles model VMamba-T achieves APbox of 46.5 and APmask of 42.1 while Vim-T achives 45.7 and 39.2 respecitively
VMamba beats Vim on ADE20k benchmark
- numbers are not directly comparable but Vmamaba seems to have better perfomance
Arxiv 24.01.24 : SegMamba: Long-range Sequential Modeling Mamba For 3D Medical Image Segmentation
#segmentation #computervision
This paper integrates the Mamba model with a U-shape structure for 3D medical image segmentation, aiming to efficiently process high-dimensional images.
What’s New: SegMamba’s combines the Mamba model, known for handling long-range dependencies, with a U-shaped architecture, with application in 3D medical image segmentation.
How it Works: SegMamba employs a Mamba encoder, a 3D decoder, and skip-connections. The Mamba encoder, with depth-wise convolution and flattening operations, reduces computational load while handling high-dimensional features.
Results: On the BraTS2023 dataset, SegMamba achieved best performance.
Why it Matters: SegMamba’s is quite niche but confims again universality of Mamba block.
We’re Thinking: Questions arise about SegMamba’s applicability to various medical imaging forms and datasets, and its comparison with other state-of-the-art methods.
In Detail
SegMamba vs Umamba
The key difference is in their approach to enhancing the U-Net architecture. SegMamba emphasizes the Mamba model for 3D segmentation, while U-Mamba combines CNNs with SSMs for versatile segmentation tasks.
Arxiv 24.01.24: MambaByte: Token-free Selective State Space Model
#llm #tokenfree
This model is token-free, directly learning from raw bytes and bypassing the biases inherent in subword tokenization.
What’s New
- “MambaByte” is a token-free language model, a novel approach as it directly learns from raw bytes, no tokenization.
- A unique perspective on efficiency and performance compared to other byte-level models.
How It Works: “MambaByte” slightly modfies Mamba module to accept raw bytes.
Results: The model surpasses the performance of state-of-the-art subword Transformers with lower computational resources. It demonstrates linear scaling in sequence length, leading to faster inference times.
Behind the News: “MambaByte” proposes an alternative to autoregressive Transformers, libertaing us from tokenization.
Why It Matters: This efficient byte sequence processing opens new avenues for language models in large-scale and diverse applications.
We’re Thinking: This paper shows a potential for token free llm learning. New applications outside of LLM should come soon.
Arxiv 24.01.25 Vivim: a Video Vision Mamba for Medical Video Object Segmentation
#video #biomedical
A framework for medical video object segmentation, focusing on addressing challenges in long-sequence modeling in video analysis. It uses a Temporal Mamba Block, which allows the model to obtain excellent segmentation results with improved speed performance compared to existing methods.
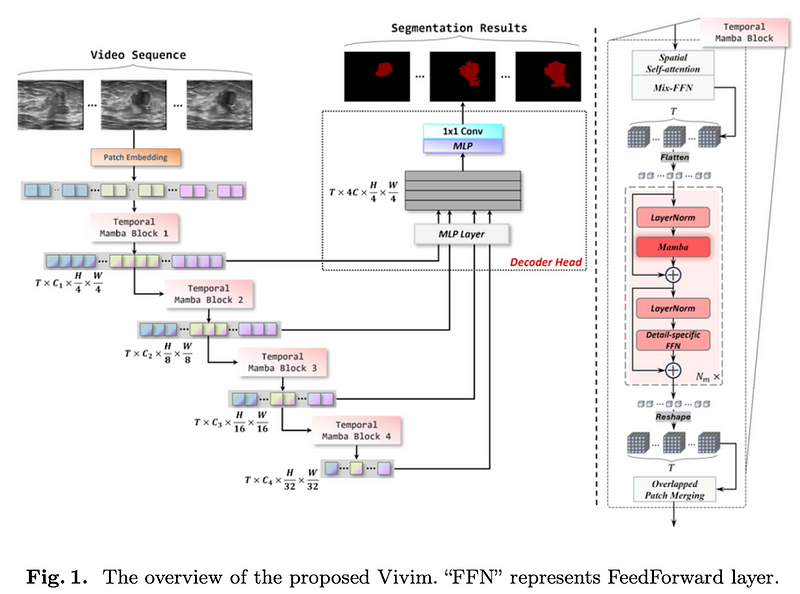
What’s New: Vivim integrates the Mamba model into a multi-level transformer architecture, transforming video clips into feature sequences containing spatiotemporal information at various scales.
How It Works
- The Temporal Mamba Block employs a sequence reduction process for efficiency, integrating a spatial self-attention module and a Mix-FeedForward layer.
- The Mamba module explores correlations among patches of input frames, while a Detail-specific FeedForward preserves fine-grained details.
- A lightweight CNN- based decoder head integrates multi-level feature sequences to predict segmentation masks.
Results: Vivim demonstrates superior performance on the breast US dataset outperforming existing video- and image-based segmentation methods.
Why It Matters: Vivim’s approach represents a significant advancement in medical video analysis, particularly for tasks like lesion segmentation in ultrasound videos.
We’re Thinking: What are the potential limitations or challenges in scaling Vivim for broader clinical applications? The paper focuses solely on breast ultrasound videos, which may limit generalizability.
In Detail:
Temporal Mamba Block
- This block starts with a spatial self-attention module for extracting spatial features, followed by a Mix-FeedForward layer.
- For temporal modeling, it transposes and flattens the spatiotemporal feature embedding into a 1D long sequence.
- The Mamba module within the block tackles the correlation among patches in input frames, while the Detail-specific FeedForward focuses on preserving fine-grained details through depth-wise convolution.
Arxiv 24.01.25: MambaMorph: a Mamba-based Backbone with Contrastive Feature Learning for Deformable MR-CT Registration
#biomedical
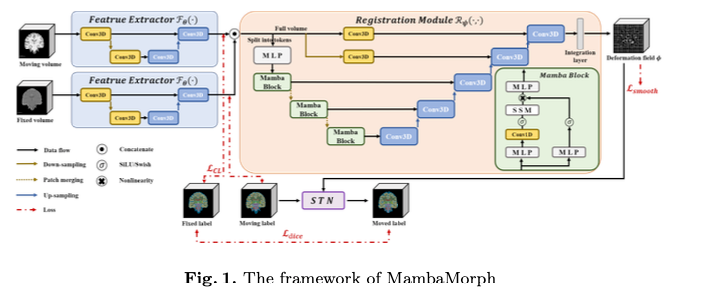
A multi-modality deformable registration network designed specifically for aligning Magnetic Resonance (MR) and Computed Tomography (CT) images.
What’s New: MambaMorph combines Mamba blocks with a feature extractor.
How It Works: MambaMorph integrates a Mamba-based backbone with contrastive feature learning for deformable MR-CT registration.
- Mamba-Based Registration Module: This module utilizes the Mamba blocks for efficient handling and processing of high-dimensional imaging data.
- Contrastive Feature Learning: a feature extractor that employs supervised contrastive learning. This is designed to learn fine-grained, modality-specific features from the MR and CT images.
Results: MambaMorph demonstrates superior performance over existing methods in MR-CT registration, showing improvements in accuracy and efficiency.
Behind the News: The development of MambaMorph is a significant step in addressing the prevalent issues in multi-modality image registration, particularly in the context of MR and CT images.
Why It Matters: The success of MambaMorph in MR-CT image registration has significant implications for medical imaging analysis.
We’re Thinking: While MambaMorph shows promising results, questions remain about its applicability to other forms of medical imaging and its performance in varied clinical scenarios.